Oreochromis niloticus is a highly nutritious aquatic food with limited shelf life. The mode of preservation of O. niloticus has raised concerns about its safety and public health due to food borne illnesses. Studies have shown that chemical preservatives are toxic and harmful to humans thereby leading to growing interest in Calotropis procera for its antibacterial properties, and there is limited information on the use of C. procera silver nanoparticles (CP-AgNPs) to preserve O. niloticus. Hence, this study aimed at evaluating its preservative effect of CP-AgNPs on O. niloticus. A total of one hundred and fifty-six samples of adult wild O. niloticus (97.41± 0.95 g) were sourced from a local river using simple random sampling. The O. niloticus fishes were subjected to four different treatments which included: dipping into sterile distilled water, dipping into NaCl solution, dipping into Calotropis procera silver nanoparticles (CP-AgNPs) solution and injecting CP-AgNPs. These were allowed to stand for 30 min, thereafter drained and held in clean basket at ambient conditions for 48 h. Samples were taken at 4 h interval for microbiological analysis according to standard methods. The isolated bacteria were identified using 16S rRNA gene sequencing. All analysis was carried out in triplicates with statistical significance set at P<0.05. The microbial count showed that CP-AgNPs exhibited antimicrobial and antifungal activities. The best treatment for preservation was the injected CP-AgNPs. Bacteria identified were Pseudomonas aeruginosa, Streptococcus agalactiae, Escherichia coli, Enterobacter sichuanensis, Enterobacter cloacae, Staphylococcus aureus, Citrobacter portucalensis, Klebsiella pneumoniae, Klebsiella variicola, Proteus mirabilis and Aeromonas caviae. This study concluded that, injecting CP-AgNPs into O. niloticus was the best treatment option; however, CP-AgNPs displayed antibacterial activities and preservative effect on O. niloticus.
| Published in | World Journal of Food Science and Technology (Volume 8, Issue 4) |
| DOI | 10.11648/j.wjfst.20240804.15 |
| Page(s) | 115-125 |
| Creative Commons |
This is an Open Access article, distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution and reproduction in any medium or format, provided the original work is properly cited. |
| Copyright |
Copyright © The Author(s), 2024. Published by Science Publishing Group |
Antibacterial, Calotropis procera, Natural Food Preservative, Oreochromis niloticus, Silver Nanoparticles
Hrs | Distilled water (cfu/g) | NaCl (cfu/g) | CP-AgNPs dipping (cfu/g) | CP-AgNps Injecting (cfu/g) |
|---|---|---|---|---|
0 | 3.60±0.17a × 104 | 2.44±0.04b × 104 | 2.20±0.04b × 104 | 2.40±0.03b × 104 |
4 | 9.58±0.08a × 104 | 7.80±0.10b × 104 | 6.20±0.13c × 104 | 6.02±0.07c × 104 |
8 | 3.29±0.08a × 105 | 2.44±0.10b × 105 | 1.52±0.04c × 105 | 1.42±0.04c × 105 |
12 | 8.71±0.18a × 105 | 6.56±0.11b × 105 | 2.06±0.01c × 105 | 1.80±0.04c × 105 |
16 | 1.91±0.03a × 106 | 1.51±0.05b × 106 | 3.34±0.04c × 105 | 2.77±0.03c × 105 |
20 | 3.28±0.04a × 106 | 3.05±0.08b × 106 | 7.11±0.06c × 105 | 4.89±0.06d × 105 |
24 | 8.50±0.06a × 106 | 6.40±0.04b × 106 | 1.42±0.06c × 106 | 1.21±0.04d × 106 |
28 | 1.74±0.01a × 107 | 1.01±0.01b × 107 | 2.77±0.06c × 106 | 2.12±0.08d × 106 |
32 | 3.84±0.02a × 107 | 3.11±0.08b × 107 | 5.89±0.07c × 106 | 4.38±0.01d × 106 |
36 | 7.64±0.01a × 107 | 5.42±0.09b × 107 | 1.84±0.01c × 107 | 1.56±0.01d × 107 |
40 | 1.21±0.02a × 108 | 1.01±0.03b × 108 | 4.57±0.01c × 107 | 3.75±0.02d × 107 |
44 | 4.42±0.03a × 108 | 4.08±0.02b × 108 | 1.17±0.02c × 108 | 1.00±0.02d × 108 |
48 | 1.17±0.02a × 109 | 1.01±0.05b × 109 | 7.74±0.06c × 108 | 7.47±0.05d × 108 |
Hrs | Distilled water (cfu/g) | Nacl (cfu/g) | CP-AgNPs dipping (cfu/g) | CP-AgNps Injecting (cfu/g) |
|---|---|---|---|---|
0 | NIL | NIL | NIL | NIL |
4 | NIL | NIL | NIL | NIL |
8 | NIL | NIL | NIL | NIL |
12 | NIL | NIL | NIL | NIL |
16 | 1.20±0.03a × 103 | NIL | NIL | NIL |
20 | 2.80±0.04a × 103 | 2.00±0.07b × 103 | NIL | NIL |
24 | 5.10±0.10a × 103 | 3.20±0.08b × 103 | NIL | NIL |
28 | 8.30±0.08a × 103 | 6.80±0.13b × 103 | NIL | NIL |
32 | 1.02±0.01a × 104 | 8.80±0.04b × 103 | 2.40±0.10c × 103 | NIL |
36 | 1.22±0.03a × 104 | 9.60±0.11b × 103 | 3.20±0.10c × 103 | NIL |
40 | 1.57±0.01a × 104 | 1.14±0.01b × 104 | 4.00±0.10c × 103 | 3.40±0.10d × 103 |
44 | 1.74±0.06a × 104 | 1.28±0.04b × 104 | 5.60±0.16c × 103 | 4.80±0.11c × 103 |
48 | 1.98±0.11a × 104 | 1.51±0.08b × 104 | 6.20±0.13c × 103 | 5.90±0.13c × 103 |
Hrs | Distilled water (cfu/g) | Nacl (cfu/g) | CP-AgNPs dipping (cfu/g) | CP-AgNps Injecting (cfu/g) |
|---|---|---|---|---|
0 | 4.50±0.13a × 103 | 1.50±0.11b × 103 | 1.00±0.09c × 102 | NIL |
4 | 8.60±0.13a × 103 | 5.60±0.08b × 103 | 2.60±0.06c × 102 | NIL |
8 | 1.02±0.03a × 104 | 7.30±0.10b × 103 | 3.60±0.14c × 102 | 1.20±0.09d × 102 |
12 | 1.28±0.04a × 104 | 1.04±0.02b × 104 | 4.00±0.18c × 102 | 2.00±0.18d × 102 |
16 | 1.84±0.04a × 104 | 1.60±0.04b × 104 | 5.80±0.04c × 102 | 3.00±0.13 d × 102 |
20 | 2.52±0.11a × 104 | 1.88±0.08b × 104 | 7.40±0.14c × 102 | 4.80±0.13d × 102 |
24 | 2.83±0.06a × 104 | 2.34±0.10b × 104 | 9.10±0.16c × 102 | 6.60±0.13d × 102 |
28 | 3.18±0.14a × 104 | 2.62±0.04b × 104 | 1.12±0.02c × 103 | 9.20±0.13c × 102 |
32 | 7.80±0.10a × 104 | 4.01±0.05b × 104 | 1.32±0.01c × 103 | 1.11±0.02d × 103 |
36 | 1.26±0.02a × 105 | 8.02±0.11b × 104 | 2.05±0.13c × 103 | 1.88±0.03c × 103 |
40 | 1.43±0.02a × 105 | 1.00±0.01b × 105 | 3.51±0.02c × 103 | 2.78±0.11d × 103 |
44 | 1.83±0.03a × 105 | 1.40±0.02b × 105 | 5.03±0.08c × 103 | 4.31±0.09d × 103 |
48 | 2.24±0.02a × 105 | 2.15±0.03b × 105 | 8.05±0.13c × 103 | 6.78±0.13d × 103 |
Hrs | Distilled water (cfu/g) | Nacl (cfu/g) | CP-AgNPs dipping (cfu/g) | CP-AgNps Injecting (cfu/g) |
|---|---|---|---|---|
0 | 6.40±0.13a × 104 | 5.20±0.52b × 104 | 3.20±0.13c × 104 | NIL |
4 | 7.40±0.16a × 104 | 5.60±0.17b × 104 | 3.60±0.13c × 104 | NIL |
8 | 9.60±0.33a × 104 | 8.50±0.28b × 104 | 3.20±0.06c × 104 | 0.70±0.09d × 103 |
12 | 1.42±0.13a × 105 | 9.80±0.35b × 104 | 3.00±0.08c × 104 | 1.60±0.14 c × 104 |
16 | 1.64±0.07a × 105 | 1.24±0.04b × 105 | 3.80±0.09c × 104 | 2.00±0.16d × 104 |
20 | 1.95±0.08a × 105 | 1.42±0.08b × 105 | 4.20±0.16c × 104 | 2.80±0.13c × 104 |
24 | 2.10±0.18a × 105 | 1.70±0.09b × 105 | 6.60±0.13c × 104 | 3.60±0.16d × 104 |
28 | 2.28±0.14a × 105 | 1.92±0.12b × 105 | 8.40±0.42c × 104 | 5.20±0.11d × 104 |
32 | 2.53±0.08a × 105 | 2.28±0.11b × 105 | 9.60±0.18c × 104 | 6.80±0.57 d × 104 |
36 | 2.75±0.11a × 105 | 2.43±0.08b × 105 | 1.13±0.07c × 105 | 8.60±0.85d × 104 |
40 | 3.28±0.28a × 105 | 2.92±0.28a × 105 | 1.22±0.09b × 105 | 9.4±0.57b × 104 |
44 | 6.40±0.25a × 105 | 4.40±0.29b × 105 | 1.58±0.09c × 105 | 1.25±0.11c × 105 |
48 | 8.50±0.27a × 105 | 6.00±0.18b × 105 | 2.35±0.11c × 105 | 1.82±0.08d × 105 |
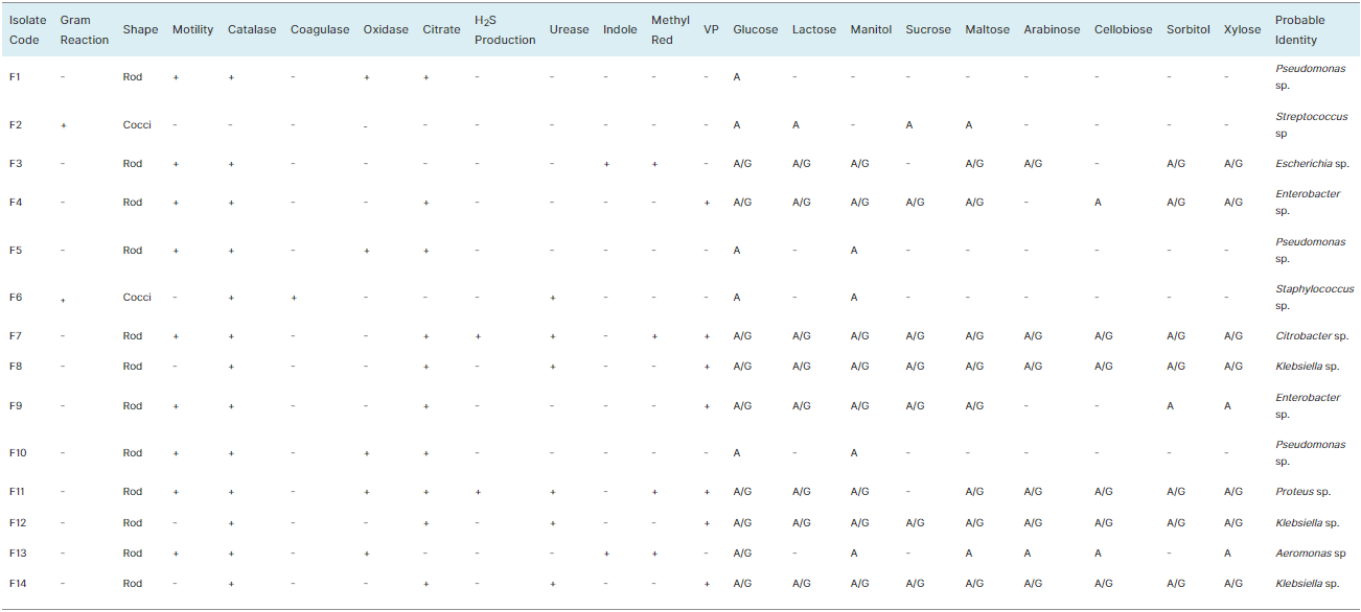
Sample ID | Predicted Organism | % Identity | Gene bank Accession No | Alignment Score | Highest Query Coverage (%) |
|---|---|---|---|---|---|
F1 | Pseudomonas aeruginosa | 98.20 | JF513146.1 | ≥200 | 99 |
F2 | Streptococcus agalactiae | 98.47 | CP050455.1 | ≥200 | 99 |
F3 | Escherichia coli | 97.82 | KF917161.1 | ≥200 | 99 |
F4 | Enterobacter sichuanensis | 99.87 | CP027986.1 | ≥200 | 99 |
F5 | Pseudomonas aeruginosa | 93.80 | MF943159.1 | ≥200 | 99 |
F6 | Staphylococcus aureus | 99.00% | CP045560.1 | ≥200 | 99 |
F7 | Citrobacter portucalensis | 94.05 | CP089316.1 | ≥200 | 99 |
F8 | Klebsiella pneumoniae | 96.81 | CP030320.1 | ≥200 | 99 |
F9 | Enterobacter cloacae | 92.58 | MZ157016.1 | ≥200 | 97 |
F10 | Pseudomonas aeruginosa | 98.14 | JF513146.1 | ≥200 | 99 |
F11 | Proteus mirabilis | 90.12 | MF977359.1 | ≥200 | 99 |
F12 | Klebsiella variicola | 97.71 | KU359261.1 | ≥200 | 99 |
F13 | Aeromonas caviae | 91.95 | OM842836.1 | ≥200 | 99 |
F14 | Klebsiella pneumoniae | 98.80 | CP121133.1 | ≥200 | 99 |
CP-AgNPs | Calotropis procera Silver Nanoparticles |
AgNPs | Silver Nanoparticles |
NPs | Nanoparticles |
AgNO3 | Silver Nitrate |
NaCl | Sodiun Chloride |
MR-VP | Methyl Red – Voges Proskauer |
DNA | Deoxyribonucleic Acid |
PCR | Polymerase Chain Reaction |
UV | Ultraviolet |
EDTA | Ethylenediaminetetraacetic Acid |
| [1] |
Diallo, I., Snoeks, J., Freyhof, J., Geelhand, D., & Hughes, A. (2020). Oreochromis niloticus The IUCN red list of threatened species. e. T166975A134879289.
https://dx.doi.org/10.2305/IUCN.UK.2020-3.RLTS.T166975A134879289.en |
| [2] | Mili, S., Ennouri, R., Fatnassi, M., Zarrouk, H., Thabet, R., & Laouar, H. (2023). Nile tilapia “Oreochromis niloticus” farming in fresh and geothermal waters in Tunisia: A comparative study. In Intensive Animal Farming - A Cost-Effective Tactic. IntechOpen. |
| [3] | Obirikorang, K. A., Opoku, E. N., Gyampoh, B. A. (2022). Feed digestion, growth and disease prevalence in Nile Tilapia (Oreochromis niloticus) cultured at different water exchange rates in a recirculating aquaculture system. Aquaculture Studies, 22(3), AQUAST565. |
| [4] | Mehmood, T., Arshad, H., Nawaz, S., Ullah, A., Hafeez, A., Anwar, F., Ahmad, M. M., Iqbal, M. (2020). Pharmaceutical potential and phenolics profiling of leaves and bark of Calotropis procera in relation to extraction solvents. Pharmaceutical Chemistry Journal, 54, 631-641. |
| [5] | Qian, Y., Yu, H., He, D., Yang, H., Wang, W., Wan, X., & Wang, L. (2013). Biosynthesis of silver nanoparticles by the endophytic fungus Epicoccum nigrum and their activity against pathogenic fungi. Bioprocess and Biosystems Engineering, 36(11), 1613-1619. |
| [6] | Alkammash, N. M. (2017). Synthesis of silver nanoparticles from Artemisia sieberi and Calotropis procera medical plant extracts and their characterization using SEM analysis. Biosciences Biotechnology Research Asia, 14(2), 521-526. |
| [7] | Kemala, P., Idroes, R., Khairan, K., Ramli, M., Jalil, Z., Idroes, G. M., Tallei, T. E., Helwani, Z., Safitri, E., Iqhrammullah, M., & Nasution, R. (2022). Green synthesis and antimicrobial activities of silver nanoparticles using Calotropis gigantea from Ie Seu- Um Geothermal area, Aceh Province, Indonesia. Molecules, 27(16), 5310. |
| [8] | Akshaya, T., Aravind, M., Manoj Kumar, S., & Divya, B. (2022). Evaluation of in-vitro antibacterial activity against gram-negative bacteria using silver nanoparticles synthesized from Dypsis lutescens leaf extract. Journal of the Chilean Chemical Society, 67(2), 5477-5483. |
| [9] | Bao, Y., He, J., Song, K., Guo, J., Zhou, X., & Liu, S. (2021). Plant-extract-mediated synthesis of metal nanoparticles. Journal of Chemistry, 2021, 1-10. |
| [10] | Nawab, R., Ali, M., Haroon, U., Kamal, A., Akbar, M., Anwar, F., Ahmed, J., Chaudhary, H. J., Iqbal, A., Hashem, M., Alamri, S., ALHaithloul, H. A. S., & Munis, M. F. H. (2022). Calotropis procera (L.) mediated synthesis of AgNPs and their application to control leaf spot of Hibiscus rosa-sinensis (L.). Brazilian Journal of Biology, 84, e261123. |
| [11] | Essien, E. R., Atasie, V. N., Udobang, E. U., & Umanu, G. (2019). Preparation of monodispersed and cytotoxic silver nanoparticles using Launaea taraxacifolia leaf extract. Journal of Nanostructure in Chemistry, 9, 259-268. |
| [12] | Dada, A. O., Ojediran, O. J., Dada, F. E., Olalekan, A. P., & Awakan, O. J. (2017). Green synthesis and characterization of silver nanoparticles using Calotropis procera extract. Journal of Applied Chemical Science International, 8(4), 137-143. |
| [13] | Harrigan, W. F., & McCance, M. E. (1976). Laboratory Methods in Microbiology. Academic Press. |
| [14] | Fawole, M. O., & Oso, B. A. (2004). Characterization of bacteria: Laboratory manual of Microbiology (4th ed.). Spectrum Book Ltd. |
| [15] | Seeley, H. W., & Van Demark, P. J. (1972). Microbes in action; a laboratory manual of Microbiology (2nd ed.). W. H. Freeman and Co. |
| [16] |
MacWilliams, M. P. (2009a). Citrate test protocol. American Society for Microbiology.
http://www.asmscience.org/content/education/protocol/protocol.3203 |
| [17] | Cheesbrough, M. (2006). District laboratory practice in tropical countries (2nd ed.). Cambridge University Press. |
| [18] | Olutiola, P. O., Famurewa, O., & Sonntag, H. G. (2000). Introduction to general Microbiology: A practical approach (2nd ed.). Bolabay Publications. |
| [19] |
MacWilliams, M. P. (2009b). Indole test protocol. American Society for Microbiology.
http://www.asmscience.org/content/education/protocol/protocol.3202 |
| [20] |
Aryal, S. (2022a). Voges-Proskauer (VP) test- principle, reagents, procedure and result. Retrieved from
https://microbiologyinfo.com/voges-proskauer-vp-test-principle-reagents-procedure-and-result/ |
| [21] |
Aryal, S. (2022b). Urease test- principle, media, procedure and result. Retrieved from
https://microbiologyinfo.com/urease-test-principle-media-procedure-and-result/ |
| [22] | Aryal, S. (2022c). Fermentation test – principle, procedure, uses and interpretation. Retrieved from |
| [23] | Lee, P. Y., Costumbrado, J., Hsu, C. Y., & Kim, Y. H. (2012). Agarose gel electrophoresis for the separation of DNA fragments. Journal of visualized experiments, (62), 3923. |
| [24] | Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95-98. |
| [25] | Salvatore, I. (2021). Food microbial quality and safety. Archives of Clinical Microbiology, 12(5), 171-172. |
| [26] | Khan, A., Rind, R., Shoaib, M., Ali, A., Mughal, G., Lakho, S. A., Malhi, K. K., Nizamani, A., & Yousaf, A. (2015). Isolation, identification and antibiogram of Escherichia coli from table eggs. Journal of Animal Health and Production, 4, 1-5. |
| [27] | De-Freitas, C. D. T., Lopes, J. L. D., Beltramini, L. M., de Oliveira, R. S. B., Oliveira, J. T. A., & Ramos, M. V. (2011). Osmotin from Calotropis procera latex: New insights into structure and antifungal properties. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1808(10), 2501-2507. |
| [28] | Ishnava, K. B., Chauhan, J. B., Garg, A. A., & Thakkar, A. M. (2012). Antibacterial and phytochemical studies on Calotropis gigantia (L.) R. Br. latex against selected cariogenic bacteria. Saudi Journal of Biological Sciences, 19(1), 87-91. |
| [29] | Mohamed, N. H., Ismail, M. A., Abdel-Mageed, W. M., & Shoreit, A. A. M. (2014). Antimicrobial activity of latex silver nanoparticles using Calotropis procera, Asian Pacific Journal of Tropical Biomedicine, 4(11), 876-883. |
| [30] | Liu, X. D., & Xu, Y. (2008). A novel raw starch digesting α-amylase from a newly isolated Bacillus sp. YX-1: Purification and characterization. Bioresource Technology, 99(10), 4315-4320. |
| [31] | Akinola, S. A., & Osundahunsi, O. F. (2017). Lactic acid bacteria and yeast diversities in spontaneously fermented millet sourdoughs. Journal of Microbiology, Biotechnology and Food Sciences, 6(4), 1030-1035. |
| [32] | Admasu, F., Mikru, A., Balkew, K., & Adane, M. (2023). Microbial profile of fresh and semicooked nile tilapia (Oreochromis niloticus) and hygienic practice of fish handlers in hawassa, Ethiopia. International Journal of Microbiology, 14, 5866719. |
| [33] | El-Tahtawi, N. F., Fudllalah, F. M. N., Alhalili, Z. A., & Aljohani, M. A. (2021). Investigation of the microbiological and physiological profile of nile tilapia "Oreochromis niloticus". Pharmacophore, 12(2), 108-115. |
| [34] | Eves, A., Turner, C., Yakupitiyage, A., Tongdee, N., & Ponza, S. (1995). The microbiological and sensory quality of septage-raised Nile tilapia (Oreochromis niloticus). Aquaculture, 132(3-4), 261-272. |
| [35] | Onjong, H. A., Ngayo, M. O., Mwaniki, M., Wambui, J., & Njage, P. M. K. (2018). Microbiological safety of fresh tilapia (Oreochromis niloticus) from Kenyan fresh water fish value chains. Journal of Food Protection, 81(12), 1973-1981. |
| [36] | Sarkar, S., Dey, S. K., Nipu, A. I., Brishti, P. S., & Billah, M. B. (2020). Microbiological assessment of Nile tilapia Oreochromis niloticus collected from different super shops and local market in Dhaka, Bangladesh. Journal of Fisheries. 8(2), 784–791. |
APA Style
Animashaun, O., Aina, D., Thonda, O. (2024). Microbiological Assessment of Oreochromis niloticus Treated with Calotropis procera - Silver Nanoparticles. World Journal of Food Science and Technology, 8(4), 115-125. https://doi.org/10.11648/j.wjfst.20240804.15
ACS Style
Animashaun, O.; Aina, D.; Thonda, O. Microbiological Assessment of Oreochromis niloticus Treated with Calotropis procera - Silver Nanoparticles. World J. Food Sci. Technol. 2024, 8(4), 115-125. doi: 10.11648/j.wjfst.20240804.15
AMA Style
Animashaun O, Aina D, Thonda O. Microbiological Assessment of Oreochromis niloticus Treated with Calotropis procera - Silver Nanoparticles. World J Food Sci Technol. 2024;8(4):115-125. doi: 10.11648/j.wjfst.20240804.15
@article{10.11648/j.wjfst.20240804.15,
author = {Oluwatoyin Animashaun and Daniel Aina and Oluwakemi Thonda},
title = {Microbiological Assessment of Oreochromis niloticus Treated with Calotropis procera - Silver Nanoparticles
},
journal = {World Journal of Food Science and Technology},
volume = {8},
number = {4},
pages = {115-125},
doi = {10.11648/j.wjfst.20240804.15},
url = {https://doi.org/10.11648/j.wjfst.20240804.15},
eprint = {https://article.sciencepublishinggroup.com/pdf/10.11648.j.wjfst.20240804.15},
abstract = {Oreochromis niloticus is a highly nutritious aquatic food with limited shelf life. The mode of preservation of O. niloticus has raised concerns about its safety and public health due to food borne illnesses. Studies have shown that chemical preservatives are toxic and harmful to humans thereby leading to growing interest in Calotropis procera for its antibacterial properties, and there is limited information on the use of C. procera silver nanoparticles (CP-AgNPs) to preserve O. niloticus. Hence, this study aimed at evaluating its preservative effect of CP-AgNPs on O. niloticus. A total of one hundred and fifty-six samples of adult wild O. niloticus (97.41± 0.95 g) were sourced from a local river using simple random sampling. The O. niloticus fishes were subjected to four different treatments which included: dipping into sterile distilled water, dipping into NaCl solution, dipping into Calotropis procera silver nanoparticles (CP-AgNPs) solution and injecting CP-AgNPs. These were allowed to stand for 30 min, thereafter drained and held in clean basket at ambient conditions for 48 h. Samples were taken at 4 h interval for microbiological analysis according to standard methods. The isolated bacteria were identified using 16S rRNA gene sequencing. All analysis was carried out in triplicates with statistical significance set at PPseudomonas aeruginosa, Streptococcus agalactiae, Escherichia coli, Enterobacter sichuanensis, Enterobacter cloacae, Staphylococcus aureus, Citrobacter portucalensis, Klebsiella pneumoniae, Klebsiella variicola, Proteus mirabilis and Aeromonas caviae. This study concluded that, injecting CP-AgNPs into O. niloticus was the best treatment option; however, CP-AgNPs displayed antibacterial activities and preservative effect on O. niloticus.
},
year = {2024}
}
TY - JOUR T1 - Microbiological Assessment of Oreochromis niloticus Treated with Calotropis procera - Silver Nanoparticles AU - Oluwatoyin Animashaun AU - Daniel Aina AU - Oluwakemi Thonda Y1 - 2024/12/12 PY - 2024 N1 - https://doi.org/10.11648/j.wjfst.20240804.15 DO - 10.11648/j.wjfst.20240804.15 T2 - World Journal of Food Science and Technology JF - World Journal of Food Science and Technology JO - World Journal of Food Science and Technology SP - 115 EP - 125 PB - Science Publishing Group SN - 2637-6024 UR - https://doi.org/10.11648/j.wjfst.20240804.15 AB - Oreochromis niloticus is a highly nutritious aquatic food with limited shelf life. The mode of preservation of O. niloticus has raised concerns about its safety and public health due to food borne illnesses. Studies have shown that chemical preservatives are toxic and harmful to humans thereby leading to growing interest in Calotropis procera for its antibacterial properties, and there is limited information on the use of C. procera silver nanoparticles (CP-AgNPs) to preserve O. niloticus. Hence, this study aimed at evaluating its preservative effect of CP-AgNPs on O. niloticus. A total of one hundred and fifty-six samples of adult wild O. niloticus (97.41± 0.95 g) were sourced from a local river using simple random sampling. The O. niloticus fishes were subjected to four different treatments which included: dipping into sterile distilled water, dipping into NaCl solution, dipping into Calotropis procera silver nanoparticles (CP-AgNPs) solution and injecting CP-AgNPs. These were allowed to stand for 30 min, thereafter drained and held in clean basket at ambient conditions for 48 h. Samples were taken at 4 h interval for microbiological analysis according to standard methods. The isolated bacteria were identified using 16S rRNA gene sequencing. All analysis was carried out in triplicates with statistical significance set at PPseudomonas aeruginosa, Streptococcus agalactiae, Escherichia coli, Enterobacter sichuanensis, Enterobacter cloacae, Staphylococcus aureus, Citrobacter portucalensis, Klebsiella pneumoniae, Klebsiella variicola, Proteus mirabilis and Aeromonas caviae. This study concluded that, injecting CP-AgNPs into O. niloticus was the best treatment option; however, CP-AgNPs displayed antibacterial activities and preservative effect on O. niloticus. VL - 8 IS - 4 ER -